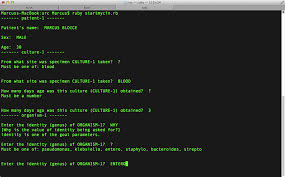
MYCIN is an
interactive expert system for infectious illness diagnosis and treatment
developed by computer scientists Edward Feigenbaum (1936–) and Bruce Buchanan
at Stanford University in the 1970s.
MYCIN was Feigenbaum's second expert system (after DENDRAL),
but it was the first to be commercially accessible as a standalone software
package.
TeKnowledge, the software business cofounded by Feigenbaum
and other partners, offered EMYCIN as the most successful expert shell for this
purpose by the 1980s.
MYCIN was developed by Feigenbaum's Heuristic Programming
Project (HPP) in collaboration with Stanford Medical School's Infectious
Diseases Group (IDG).
The expert clinical physician was IDG's Stanley Cohen.
Feigenbaum and Buchanan had read stories of antibiotics
being prescribed wrongly owing to misdiagnoses in the early 1970s.
MYCIN was created to assist a human expert in making the
best judgment possible.
MYCIN started out as a consultation tool.
MYCIN supplied a diagnosis that included the necessary
antibiotics and dose after inputting the results of a patient's blood test,
bacterial cultures, and other data.
MYCIN also served as an explanation system.
In simple English, the physician-user may ask MYCIN to
expound on a certain inference.
Finally, MYCIN had a knowledge-acquisition software that was
used to keep the system's knowledge base up to date.
Feigenbaum and his collaborators introduced two additional
features to MYCIN after gaining experience with DENDRAL.
MYCIN's inference engine comes with a rule interpreter to
begin with.
This enabled "goal-directed backward chaining" to
be used to achieve diagnostic findings (Cendrowska and Bramer 1984, 229).
MYCIN set itself the objective of discovering a useful
clinical parameter that matched the patient data submitted at each phase in the
procedure.
The inference engine looked for a set of rules that applied
to the parameter in question.
MYCIN typically required more information when evaluating
the premise of one of the rules in this parameter set.
The system's next subgoal was to get that data.
MYCIN might test out new regulations or ask the physician
for further information.
This process was repeated until MYCIN had enough data on
numerous factors to make a diagnosis.
The certainty factor was MYCIN's second unique feature.
These elements should not be seen "as conditional
probabilities, [though] they are loosely grounded on probability theory,"
according to William van Melle (then a doctoral student working on MYCIN for
his thesis project) (van Melle 1978, 314).
The execution of production rules was assigned a value
between –1 and +1 by MYCIN (dependent on how strongly the system felt about
their correctness).
MYCIN's diagnosis also included these certainty elements,
allowing the physician-user to make their own final decision.
The software package, known as EMYCIN, was released in 1976
and comprised an inference engine, user interface, and short-term memory.
It didn't have any information.
("E" stood for "Empty" at first, then
"Essential.") Customers of EMYCIN were required to link their own
knowledge base to the system.
Faced with high demand for EMYCIN packages and high interest
in MOLGEN (Feigenbaum's third expert system), HPP decided to form IntelliCorp
and TeKnowledge, the first two expert system firms.
TeKnowledge was eventually founded by a group of roughly
twenty individuals, including all of the previous HPP students who had
developed expert systems.
EMYCIN was and continues to be their most popular product.
Find Jai on Twitter | LinkedIn | Instagram
You may also want to read more about Artificial Intelligence here.
See also:
Expert Systems; Knowledge Engineering
References & Further Reading:
Cendrowska, J., and M. A. Bramer. 1984. “A Rational Reconstruction of the MYCIN Consultation System.” International Journal of Man-Machine Studies 20 (March): 229–317.
Crevier, Daniel. 1993. AI: Tumultuous History of the Search for Artificial Intelligence. Princeton, NJ: Princeton University Press.
Feigenbaum, Edward. 2000. “Oral History.” Charles Babbage Institute, October 13, 2000. van Melle, William. 1978. “MYCIN: A Knowledge-based Consultation Program for Infectious Disease Diagnosis.” International Journal of Man-Machine Studies 10 (May): 313–22.